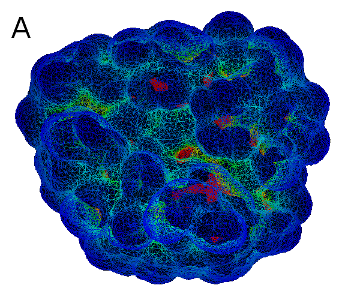
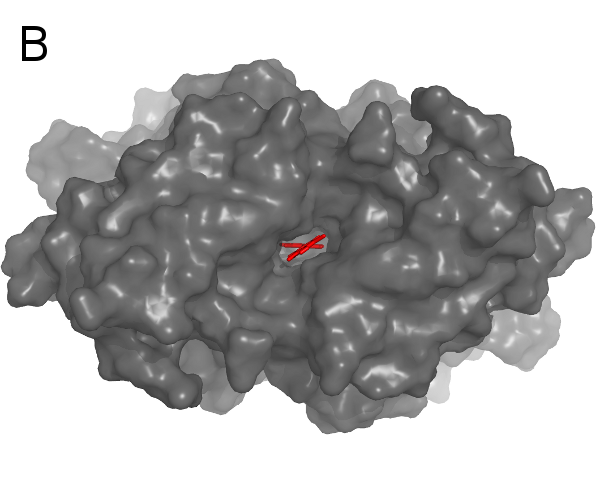
Introduction:
Cyscore is an empirical scoring function for accurate protein-ligand binding affinty prediction. It is compromised of hydrophobic free energy, van der Waals interaction energy, Hydrogen-bond energy and the ligand's entropy. To improve the prediction accuracy, a novel curvature weighted surface area model was developed for the hydrophobic free energy calculation. Our analyses show that the new model is superior to the conventional surface area model, implying that surface shape is also very important other than surface area for the prediction of hydrophobic free energy. Cyscore could be a useful tool for the accurate protein-ligand binding affinity prediction.
Please cite "Improved protein-ligand binding affinity prediction by using a curvature dependent surface area model. Yang Cao and Lei Li, Bioinformatics, 30(12):1674-1680, 2014"
Cyscore is free for non-commercial users. Other users please contact cy_scu@yeah.net .
Cyscore is a command-line application under Linux-x86. The usage is :
Cyscore [Protein PDB file] [Ligand mol2 file]
Please note: All hydrogens of proteins and ligands are needed for correct calculation.
A: a curvature-colored surface area model, B: an example of Cyscore application (PDB code: 2G5U)